Read Classification
The classification step uses ClassifyReads, a high-performance implementation of the Ribosomal Database Project (RDP) Classifier described in Wang Q. et al., 2007 (dx.doi.org/10.1128%2FAEM.00062-07). The original RDP classifier algorithm used 8-base words due to implementation constraints. ClassifyReads uses more efficient data structures and is able to use 32-base words – giving each word more specificity for each species. Below, we provide information on the classification steps used in ClassifyReads.
Word-specific Priors
Let W = { w1, w2, …, wd } be the set of all possible 32-base words. From the corpus of N sequences, let n(wi) be the number of sequences containing subsequence wi. The word-specific prior estimate of the likelihood of observing word wi in a sequence over the entire corpus can be calculated with the following formula:
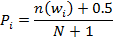
Taxonomic Conditional Probabilities
In the original RDP classifier, classifications were performed to the genus level. In ClassifyReads, we have extended this behavior to the species level. In the sections below, we denote T to represent classifications at a given taxonomic level (genus, species, or subtype). By default, we classify at the species level.
For species T with a training set consisting of M sequences, let m(wi) be the number of sequences containing word wi. The conditional probability that a member of T contains wi was estimated with the equation:
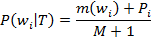
The joint probability of observing from species T a partial sequence, S, containing a set of words, V = { v1, v2, …, vf } (V ⊆ W), was estimated as:
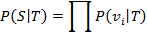
Naïve Bayesian Assignment
By Bayes’ theorem, the probability that an unknown query sequence, S, is a member of species T is:
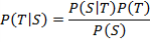
P(T) is the prior probability of a sequence being a member of T
P(S) is the overall probability of observing sequence S (from any species).
Assuming all species are equally probable (equal priors), the constant terms P(T) and P(S) can be ignored. We classify the sequence as a member of the genus giving the highest probability score.
Example
Here are the results from a typical classification (results are in log space):
Methanocaldococcaceae;MethanocaldococcusP(T|S) = -367.7
Methanococcaceae;MethanothermococcusP(T|S) = -2936
Thermofilaceaen;ThermofilumP(T|S) = -2963.2
The natural log-transformed probabilities are small due to product multiplication during the joint probability calculation. The huge difference between the best hit and the second best hit is notable. A difference of 3 would indicate 20× higher probability for the best hit. Here the difference is 2568.3 (i.e. 2.5 × 101115 higher probability).
Removing Taxonomic Levels
If the difference between the two best hits is small: 0.05 in log space (within 1.6× higher probability), we prune the taxonomic levels of the best hit until it is the same as the second best hit.
For example, consider the following results:
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;melaninogenica - P(T|S) = -200.00
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;histicola - P(T|S) = -200.04
In this case, the difference between the top two hits is less than 0.05. Therefore we prune the away the taxonomic levels from the bottom (species) to the top (kingdom) until both hits are identical. In this case, we stop pruning when we reach the Prevotella (genus) classification.
The pruning procedure reflects that we have strong evidence that the read comes from the Prevotella genus, but that we do not have enough evidence to deduce which species.
The removal threshold (0.05) was determined by analyzing misclassifications during loop-back classification of the Greengenes database.
Gaining Species Level Classification
ClassifyReads uses the full Greengenes database. Some of the entries in that database are only classified down to the genus level, while other entries are classified down to the species and subtype level. In some instances, the top results can look like this example:
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella; - P(T|S) = -200.000
Bacteria;Bacteroidetes;Bacteroidia;Bacteroidales;Prevotellaceae;Prevotella;histicola - P(T|S) = -200.000
Bacteria;Firmicutes;Bacilli;Gemellales;Gemellaceae;Gemella;haemolysans - P(T|S) = -1846
In this case, the best hit is classified down to the genus level and the second best hit is classified down to the species level. If the difference between the two best hits is near identical (difference less than 0.001 in log space), we use the species level classification instead.
Classification Confidence
The RDP classifier used a bootstrapping method of randomly subsampling the words in the sequence to determine the classification confidence.
In ClassifyReads, we no longer perform this bootstrapping procedure. This change is primarily due to performance reasons (bootstrapping slowed the algorithm down by 20–50×) and weak correlation between the resulting confidence estimate and the actual classification accuracy.
At the moment, confidence is statically assigned based on the overall accuracy of our classification algorithm at different taxonomic levels:
|
Taxonomic Level |
Accuracy |
|---|---|
|
Kingdom |
100% |
|
Phylum |
100% |
|
Class |
100% |
|
Order |
99.98% |
|
Family |
99.97% |
|
Genus |
99.65% |
|
Species |
98.24% |