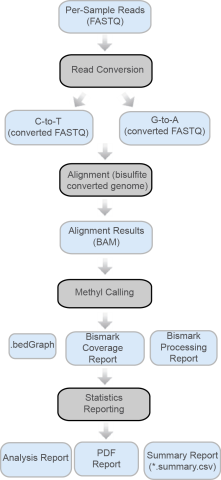
Introduction
The BaseSpace® app MethylSeq v1.0 analyzes DNA that has been sequenced using the TruSeq DNA Methylation Kit. Sequencing-based DNA methylation analysis applies the coverage density and flexibility enabled by next-generation sequencing to enhance epigenetic studies.
The process of bisulfite treatment denatures genomic DNA into single-stranded DNA (ssDNA). The TruSeq DNA Methylation Kit converts bisulfite-treated, ssDNA into an Illumina sequencing library. All ssDNA fragments are captured during the library prep procedure, eliminating sample loss associated with other methods.
The core algorithm used in MethylSeq v1.0 is Bismark, which maps bisulfite-treated sequencing reads to the genome of interest and performs methylation calls.
The alignment method used in MethylSeq v1.0 is Bowtie2. Bowtie2 is an ultrafast and memory-efficient tool for aligning sequencing reads to long reference sequences.
The MethylSeq v1.0 app generates the following output files:
| • | BAM files, which contain the reads after alignment. |
| • | Bismark Processing Report, which contains a processing report generated by Bismark. |
| • | Cytosine Report (optional), which contains methylation status for every cytosine in the genome, including both strands. |
| • | bedGraph, which contains a cytosine methylation status report for only the cytosines that have sequencing coverage. |
A summary page and other reports are available in multiple formats.
See MethylSeq v1.0 Output and MethylSeq Methods.
Figure 1 MethylSeq v1.0 Analysis with Bismark