Filter BAM Input
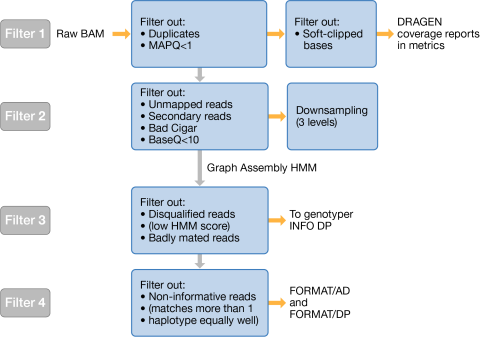
Information in the IGV BAM pileup differs from the INFO/DP and FORMAT/DP in the VCF/gVCF as a result of filtering steps applied throughout variant calling. There are four filters used to exclude reads from the genotyping calculations. The following figure summarizes the four filters.
|
•
|
Filter 1 filters out the following reads from the IGV BAM input: |
|
–
|
Soft-clipped bases. DRAGEN filters out soft-clipped bases only when calculating coverage reports. |
|
–
|
[Somatic] Reads with MAPQ < vc-min-tumor-read-qual, where vc-min-tumor-read-qual >1. |
|
•
|
Filter 2 trims bases with BQ < 10 and filters out the following reads: |
|
•
|
Filter 3 occurs after down-sampling and HMM. Filter 3 filters out the following reads: |
|
–
|
Reads that are badly mated. A badly mated read is a read where the pair is mapped to two different reference contigs. |
|
–
|
Disqualified reads. Reads are disqualified if their HMM score is below a threshold. |
|
•
|
Filter 4 occurs after the genotyper runs. The genotyper adds annotation information to the FORMAT field. Filter 4 filters out reads that are not informative. For example, if the HMM scores of the read against two different haplotypes are almost equal, the read is filtered out because it does not provide enough information to distinguish which of the two haplotypes are more likely. |
INFO/DP includes both informative and non-informative reads.
FORMAT/AD and FORMAT/DP include only informative reads.

