Calling Model
Chromosomal mosaicism is detected when there is a significant shift in median coverage of a chromosome compared to the overall autosomal median coverage.
The following table displays some examples of expected shifts in coverage for a give aneuploidy and mosaic fraction.
|
Neutral Copy Number |
Variant Copy Number |
Mosaic Fraction |
Expected Coverage Shift |
|---|---|---|---|
|
2 |
1 |
10 % |
-5% |
|
2 |
1 |
5 % |
-2.5% |
|
2 |
3 |
5 % |
+2.5% |
|
2 |
3 |
10 % |
+5% |
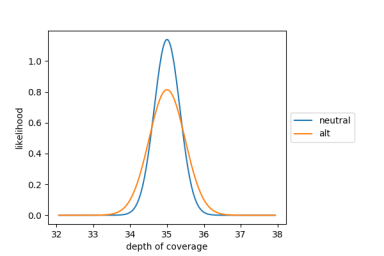
The Ploidy Caller models coverage as a normal distribution for both the null (neutral) and the alternative (mosaic) hypotheses. The two normal distributions have equal mean at the median autosomal coverage for the sample, but the variance of the alternative normal distribution is greater than that of the null normal distribution. The baseline variance of the two models at 30x coverage was determined empirically from a cohort of ~2500 WGS samples. The actual variance used for the two models is calculated from the baseline variance at 30x coverage, adjusting for the median autosomal coverage of the sample. Below are the likelihood distributions for the null and alternative hypotheses for a sample with 35x median autosomal coverage.
Null and Alternative Hypotheses Likelihood Distributions
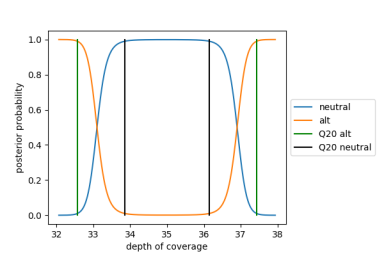
After apply an empirically estimated prior for chromosomal mosaicism, the Ploidy Caller generates ploidy calls according to the posterior probability of null and alternative hypotheses as shown below for a sample with 35x median autosomal sequencing coverage.
Null and Alternative Hypotheses Posterior Probability
A 35x median autosomal coverage, the threshold for deciding between a neutral (REF) and an alternative (DEL or DUP) call is roughly at +/- 5% shift in coverage for an autosome. A 100x median autosomal coverage, the threshold is at roughly +/0 3% shift in coverage for an autosome. A Q20 threshold is used to filter low quality calls.