Mode of Inheritance Logic
The following descriptions list the logic used for filtering by mode of inheritance.
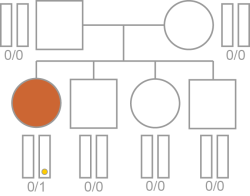
The variant is not inherited from either parent, regardless of affected status across the pedigree.
De novo inheritance is typically a consideration when both parents are unaffected. If affected siblings are in the pedigree then de novo inheritance is less likely, however the possibility cannot be completely eliminated.
Inheritance is calculated based on the following requirements.
| • | The variant is present in the proband. |
| • | Both parents are present in the pedigree. |
| • | The variant is not present in either parent. |
Typical De Novo Mutation Logic
-
 Unaffected
Unaffected -
 Affected
Affected -
 Variant
Variant
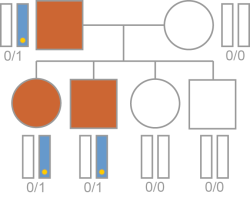
The variant is shared between the proband and all affected parents. In autosomal dominant inheritance, the variant is on an autosomal chromosome.
If both the parent and proband are affected, a dominant inheritance pattern is typical.
Inheritance is calculated based on the following requirements.
| • | The variant is present in the proband. |
| • | At least one parent is affected. |
| • | The variant is present in all affected parents. |
| • | The variant has a computed inheritance. See Inheritance Labels. |
Typical Autosomal Dominant Inheritance Logic
-
 Unaffected
Unaffected -
 Affected
Affected -
 Variant
Variant
The variant is present in the proband.
This filter can be combined with subfilters such as Not in Unaffected (NU) and In Affected (IA).
Recessive inheritance includes Recessive Singlet (RSG) and Recessive Compound Het (RCH).
Recessive inheritance is typically a consideration when both parents are unaffected because carriers do not manifest the disease phenotype in a recessive context.
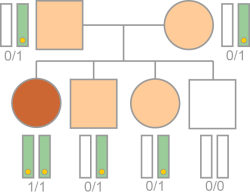
The variant follows a hemizygous or homozygous recessive pattern in the proband. In autosomal recessive inheritance, the variant is on an autosomal chromosome.
Inheritance is calculated based on the following requirements.
| • | The variant is homozygous or hemizygous in the proband. |
| • | The inheritance label is anything other than Conflict or null. |
Typical Autosomal Recessive (Homozygous) Inheritance Logic
-
 Unaffected
Unaffected -
 Unaffected carrier
Unaffected carrier -
 Affected
Affected -
 Variant
Variant
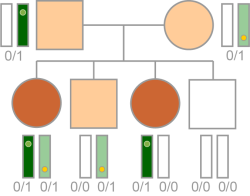
The variant follows a compound homozygous recessive pattern in the proband. Inheritance is calculated based on the following requirements.
| • | The variant is heterozygous or in the proband. |
| • | There are at least two variants in the variant set impacting the same transcript that meet transcript-level filters. |
| • | At least one of the variants in the computed inheritance set has an inheritance from both parents (Mother, Father) or at least one of Ambiguous, Conflict, Unknown, Mother/Unknown, Father/Unknown, or null. |
Typical Autosomal Recessive Inheritance (Compound Heterozygous) Logic
-
 Unaffected
Unaffected -
 Unaffected carrier
Unaffected carrier -
 Affected
Affected -
 Variant 1
Variant 1 -
 Variant 2
Variant 2
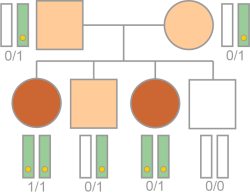
The variant follows a recessive (RSG or RCH) inheritance pattern and meets the following additional requirements.
| • | In RSG variants, the variant is homozygous or hemizygous in all affected individuals. |
| • | In RCH variants, the variant is heterozygous or hemizygous diploid loss in all affected individuals. |
If multiple siblings are affected, it is likely that both siblings have the same causal genotype. Filtering for variants where all affected individuals share the same genotype is more strict than the Shared Variant in Affected filter option, which only requires a shared variant.
Same Genotype in Affected Individuals
-
 Unaffected
Unaffected -
 Unaffected carrier
Unaffected carrier -
 Affected
Affected -
 Variant
Variant
The variant follows a recessive (RSG or RCH) inheritance pattern and meets the following additional requirements.
| • | In RSG variants, the variant is present in all affected individuals. |
| • | In RCH variants, at least one variant from a given variant set for a transcript is present in every affected individual. |
If more than two affected siblings (including proband) are in a pedigree, the affected individuals are likely to share at least one causal variant. Affected individuals are also likely to share the same genotype, however the shared variant filter option only requires shared variants. The Same Genotype in Affected filter option is more strict and requires the same genotype.
Shared Variant in Affected Individuals

-
 Unaffected
Unaffected -
 Unaffected carrier
Unaffected carrier -
 Affected
Affected -
 Variant 1
Variant 1 -
 Variant 2
Variant 2
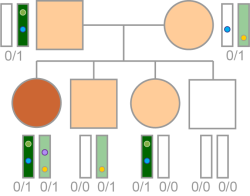
The variant follows an ambiguous recessive compound het inheritance pattern and de novo variants are absent.
If complete penetrance is assumed, variants that are ambiguously inherited can be ruled out as causal because they cannot form unique compound hets (ie, compound hets that do not exist in an unaffected parent) unless they are combined with a de novo variant.
Recessive Compound Het With De Novo Variant
-
 Unaffected
Unaffected -
 Unaffected carrier
Unaffected carrier -
 Affected
Affected -
 Variant 1
Variant 1 -
 Variant 2
Variant 2 -
 Variant 3
Variant 3 -
 De novo variant
De novo variant
Inheritance logic uses the following labels.
|
Label |
Description |
|---|---|
|
Both |
Homozygous variants that are inherited from both parents. |
|
Ambiguous |
Heterozygous variants that could have been inherited by either parent. |
|
Mother |
Zygosity is consistent with inheritance from mother. |
|
Father |
Zygosity is consistent with inheritance from father. |
|
Unknown |
Inheritance is unclear due to missing molecular data from a parent. The variant can be inherited or de novo. |
|
Mother/Unknown |
Inheritance could be from mother but is unclear due to missing molecular data from the father. |
|
Father/Unknown |
Inheritance could be from father but is unclear due to missing molecular data from the mother. |
|
Conflict |
Inheritance pattern is not consistent with typical Mendelian inheritance (eg, unclear inheritance of variant or reference alleles). |
|
Mother/Conflict |
Inheritance could be from mother but inheritance also violates expected Mendelian inheritance. |
|
Father/Conflict |
Inheritance could be from father but inheritance also violates expected Mendelian inheritance. |
The following inheritance labels appear in the MOI column of the variant grid.
| • | DN—De novo variant. |
| • | DN_LC—De novo (low confidence) |
| • | DN—Dominant inheritance. |
| • | IA—In affected. |
| • | NOAMBIG—Inheritance is not ambiguous. |
| • | NOHOM—No homozygous variants observed within the pedigree. |
| • | NU—Not in unaffected. The variant is not observed in unaffected subjects. |
| • | RCH—Recessive compound het variant. |
| • | RSG—Recessive singlet variant. |