OncoCNV Analysis Methods
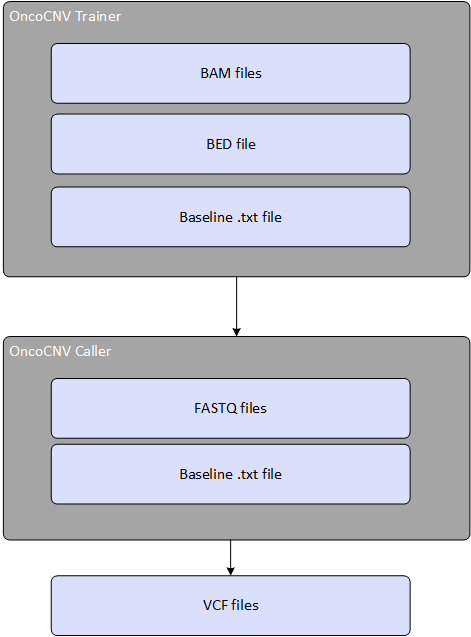
The DNA + RNA Amplicon App detects copy number variants based on circular binary segmentation and statistical tests, using normalized DNA amplicon data. The app takes BAM files and a corresponding baseline file as input and calculates CNV statistics. The baseline file must be generated by the OncoCNV trainer app.
For amplicon panel design, Illumina recommends the amplicons have neutral GC-content (eg, 0.3–0.7) and normal size (eg, < 150 bp) as GC-content, amplicon size, and other genomic content can affect coverage. In general, multiple amplicons should be designed for a targeted gene or region to reliably detect a CNV event. Illumina recommends no fewer than seven amplicons for a targeted gene or region. The normalization algorithm typically performs better for larger panels (more data points) than smaller panels, and thus larger panel may have better performance.
For more information, see Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data
OncoCNV Workflow