How to set up a PhiX validation run on the NextSeq™ 500/550 system using BaseSpace Hub Prep tab
11/12/20
A PhiX validation run confirms proper hardware and software performance of the instrument. The Illumina PhiX control library is a well-balanced genome with relatively equal representation of A, T, G, and C nucleotides. PhiX lacks an index and is not an appropriate tool for assessing Index Read performance. Below are instructions for setting up a PhiX validation run for the NextSeq™ 500/550 system using Prep tab available through BaseSpace Hub. BaseSpace Prep Tab is not available on NextSeq instruments running NextSeq Control Software v4.0 or higher. Follow instructions in the Local Run Manager: How to set up a PhiX validation run support bulletin to set up a PhiX validation run in NextSeq Control Software v4 or higher.
- Navigate to BaseSpace Hub and log in to your BaseSpace Hub account.
- Once in BaseSpace Hub, select PREP.

- Select Manual Prep - Biological Samples.
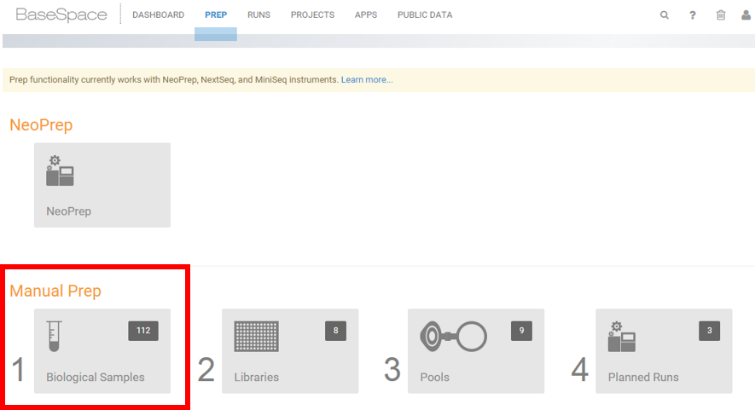
-
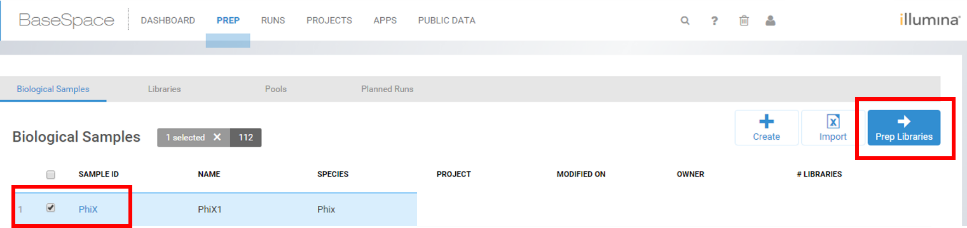
- If you already have setup a PhiX sample, select the check next to PhiX and then select Prep Libraries.
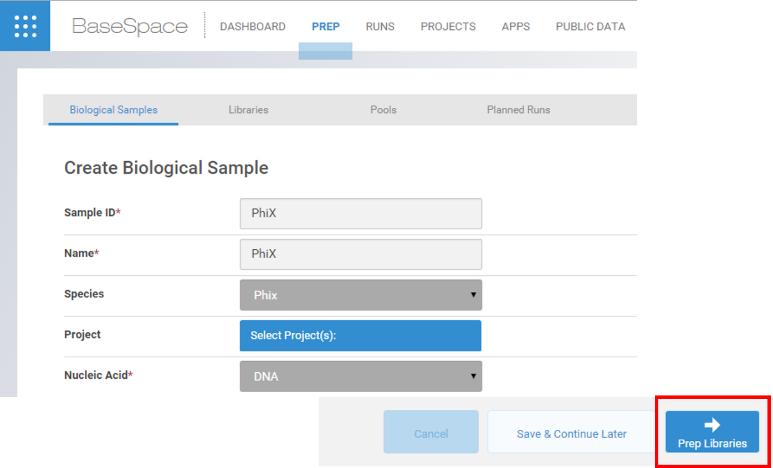
- If you do not have a PhiX sample created, select Create and then fill in the settings as below.
For Project field, select New and then fill Name with PhiX. Select Confirm for the project and then Prep Libraries.
- If you already have setup a PhiX sample, select the check next to PhiX and then select Prep Libraries.
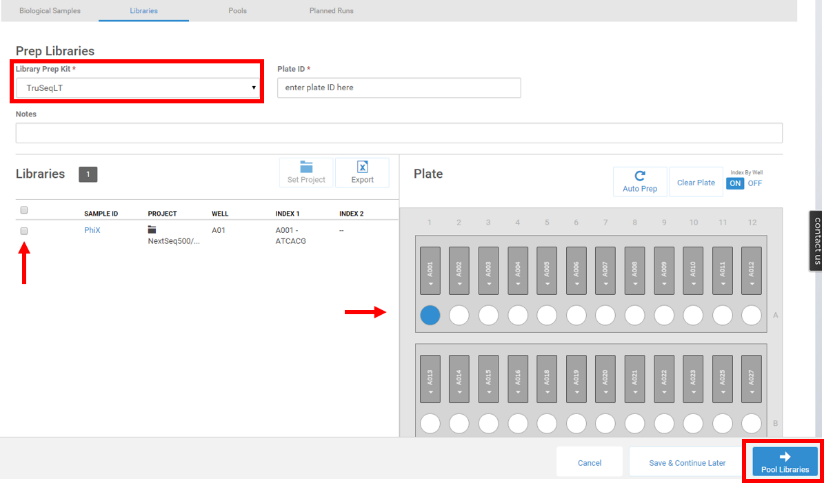
- Select TruSeq LT for Library Prep Kit and enter a Plate ID. Select your library and drag and drop into the first well of the plate. Select Pool Libraries to continue.
- Select your library from the plate on the left and drag and drop into the pool selection. Label your pool PhiX and select Plan Run.
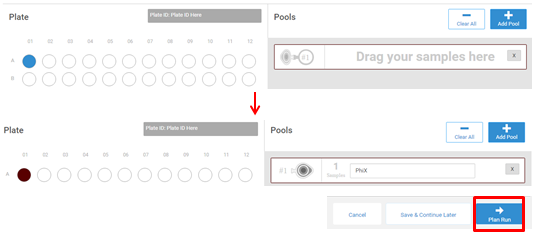
- Plan your run. Run length and parameters vary depending on troubleshooting needs and reagent chemistry version. An example setup for a 150 x150 PhiX run is shown below.
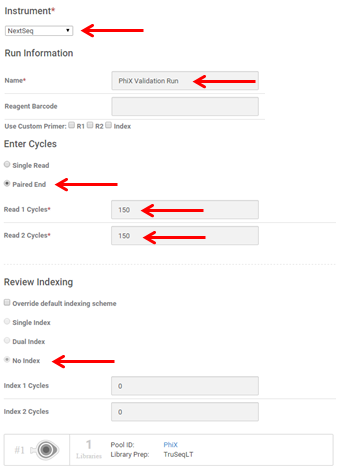
- Select Sequence to save your planned run.
For more information about BaseSpace Hub Prep tab and preparing PhiX for the NextSeq 500/550 system, see the following documents:






